How to publish biodiversity data through GBIF.org
Overview
Teaching: 30 min
Exercises: 30 minQuestions
What is IPT for the GBIF node
How is IPT organized
Objectives
Understand how IPT works.
GBIF—the Global Biodiversity Information Facility—is an international network and data infrastructure funded by the world’s governments and aimed at providing anyone, anywhere, open access to data about all types of life on Earth.
Presentation: Data Publication workflow ‘generic’
GBIF supports publication, discovery and use of four classes of data:
- Resource metadata
- Checklist Data
- Occurrence Data
- Sampling Event Data
At the simplest, GBIF enables sharing information describing a biodiversity data resource – even when no further digital information is currently available from the resource. Other data classes support an increasingly richer and wider range of information on species, their distributions and abundance.
Data publishers are strongly encouraged to share their data using the richest appropriate data class. This maximizes the usefulness of the data for users.
To give yourself an introduction to how the IPT can be used to publish biodiversity data through GBIF.org, it’s highly recommended watching this concise 25 minute live demo below:
Prerequisites
You require an account on a GBIF Integrated Publishing Toolkit (IPT) to publish your data.
Hint: it is highly recommended that you save yourself time and money by requesting an account on an IPT located at a data hosting centre in your country or community.
Hint: you could install and maintain your own IPT instance if you have technical skills and capacity to maintain it online near 100% of the time.
Hint: if no data hosting centre exists in your country, and you or your organization don’t have the technical skills and capacity to host an IPT, you can contact the GBIF Helpdesk helpdesk@gbif.org for assistance.
Assuming that you would like to register your dataset with GBIF and make it globally discoverable via GBIF.org, your dataset must be affiliated with an organization that is registered with GBIF.
Hint: to register your organization with GBIF, start by completing this online questionnaire. The registration process can take days, so in parallel you can proceed to publish your data.
Hint: if you aren’t affiliated with any organization, you can contact the GBIF Helpdesk helpdesk@gbif.org for assistance. In the meantime, you can proceed to publish your data.
Instructions
To publish your data, follow the 7 steps below.
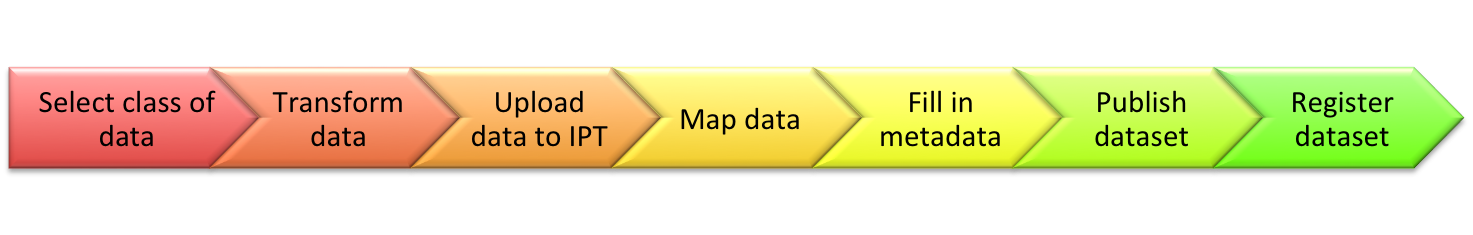
1. Select the class of biodiversity data you have from this list:
- Resource metadata
- Checklist Data
- Occurrence Data
- Sampling Event Data
2. Transform your data into a table structure, using Darwin Core (DwC) terms as column names
Hint: try using an Excel template to structure your data, and understand what DwC terms are required and recommended (Excel templates for each dataset class are available in the above links - see the previous point)
Hint: it is possible to use data stored in a supported database
3. Upload your data to the IPT
Hint: refer to other sections of this manual for additional guidance, such as the Manage Resources Menu section.
4. Map the data (e.g. Checklist Data gets mapped to the Taxon Core, Occurrence Data gets mapped to the Occurrence Core, Sampling Event Data gets mapped to the Event Core).
5. Fill in resource metadata using the IPT’s metadata editor
6. Publish the dataset (make it freely and openly available worldwide)
7. Register the dataset with GBIF.
Your organization must be registered with GBIF (see prerequisite 2 above) and added to your IPT by the IPT administrator. Otherwise, the organization will not be available to choose from in the IPT.
Exercises 1: Publish this occurrence dataset (dwc-a) on the Croatian IPT ipt.bioportal.hr
Most of the work on the publication of the data lies in the data cleaning, mapping and the description of the dataset. Once a Darwin Core archive was generated, it is fairly simple to publish it again, on another IPT for example.
Publish this dataset, “already published by the Croatian Faculty of science (which is already a GBIF data publisher) on the GBIF ECA Cloud IPT” again on the Croatian IPT. Make sure you are logged in on the IPT instance.
You should have recieved a pswdr and a login to the Croatian IPT instance.Solution
- donwload the dwc-a file here
- Go to the tab
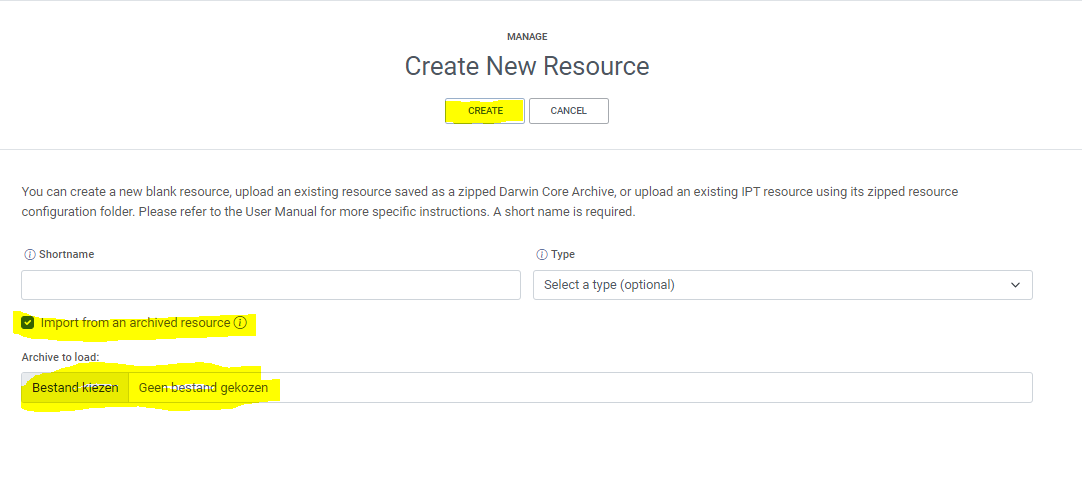
manage resources- create a new dataset
Create new dataset- provide a new shortname
- Choose
Import from an archived resource- Choose the Dwc-a file
- Click
save- If everything went correct, your metadata and data is correctly mapped in the IPT and ready to publih.
- Click
publishto finish this exercise
Exercises 2: Publish this occurrence dataset on the Croatian IPT ipt.bioportal.hr
Unfortunately, in most cases you will not have a DwC-a file availble, meaning, that you should, together with the data researcher or person who would like to publish his or her data to GBIF, create a dwc-a.
The IPT is a good tool to create dwc-archives. (There are also other tools available here for example but we do not recommend this.
For this exercise we prepared all the files needed to generate a dwc-a. Make sure you are logged in on the IPT instance.
You should have recieved a pswdr and a login to the Croatian IPT instance.
You can find an occurrence file here
You can find the metadata here Copy paste only the minimal set of information on the right place in the IPTSolution
- donwload the dwc-a file
- go to the tab
manage resources- create a new dataset
Create new dataset- provide a new shortname
- select type
occurrenceand pushcreate- deal with
source data,darwin core mappingsandmetadata(tip see session metadata & data validation)- publish your dataset
- change visibility to
public- register your dataset (not needed in this exercise)
- Click
publishto finish this exercise
Exercises 3: Publish this sample based dataset dataset on the Croatian IPT ipt.bioportal.hr
Unfortunately, in most cases you will not have a DwC-a file availble, meaning, that you should, together with the data researcher or person who would like to publish his or her data to GBIF, create a dwc-a.
The IPT is a good tool to create dwc-archives. For this exercise we prepared all the files needed to generate a dwc-a. Make sure you are logged in on the IPT instance.
You should have recieved a pswdr and a login to the Croatian IPT instance.
You can find an occurrence file here occurrence
You can find the event file here event
You can find the metadata here Copy paste only the minimal set of information on the right place in the IPTSolution
- go to the tab
manage resources- create a new dataset
Create new dataset- provide a new shortname
- select type
sampling eventand pushcreate- deal with
source dataadd both files to the IPT- deal with
darwin core mappingsfor the occurrence file- deal with
darwin core mappingsfor the event file- deal with
metadataalso here, only copy paste the minimum needed- publish your dataset
- change visibility to
public- register your dataset (not needed in this exercise)
- Click
publishto finish this exercise
Exercises 4: Publish this checklist dataset dataset on the Croatian IPT ipt.bioportal.hr
Now, we will publish a checklist data on on the IPT. A checklist is a 3rd type of dataset you can publish on Global Biodiversity Information Facility. A cheklist has no occurrences as the core file, but the species (the taxon) is at the centre of the star scheme. For this exercise we prepared all the files needed to generate a dwc-a. Make sure you are logged in on the IPT instance.
You should have recieved a pswdr and a login to the Croatian IPT instance.
You can find all the needed data here: TrIAS The TrIAS cheklist is a live ‘cheklist’ which is regurlaly updated throughGithub actionsand an automatic update function in the IPT.
You can donwload the needed files from Github. If you want to make sure your published datsets is always up to date, you can use the raw online files as a source file raw Github content For this checklist, we have a taxon, description, distribution, speciesprofile and references file. Only use (download) the taxon, description and spieciesprofile file for this exercise. You can find the metadata here Copy paste only the minimal set of information on the right place in the IPTSolution
- go to the tab
manage resources- create a new dataset
Create new dataset- provide a new shortname
- select type
checklistand pushcreate- deal with the
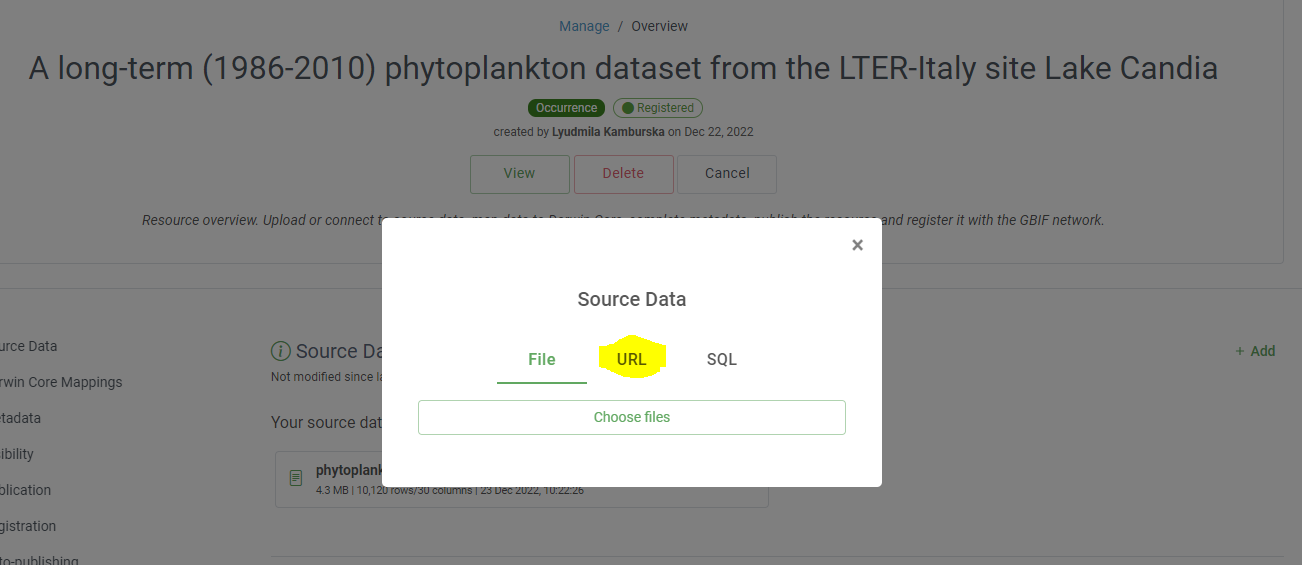
source dataimport all files in the IPT. In the IPT, for taxon choosesource data is urlinstead of file and use this url raw Github content- deal with
darwin core mappingsfor thedistributionfile- deal with
darwin core mappingsfor thedistributionfile- deal with
darwin core mappingsfor theprofilefile- deal with
metadataalso here, only copy paste the minimum needed- publish your dataset
- change visibility to
public- register your dataset (not needed in this exercise)
- Click
publishto finish this exercise
Key Points
IPT is the main tool to publish your data to GBIF